Fugue Devlog 26: More tool building and 3d nightmares.
It's been a while since the last update; I was away at a language immersion program and then very busy with our move to Paris but managed to find some time here and there to continue work on the Fugue. Though it's probably more accurate to say the "pre-work" on the game, as I'm still deep in setting up all the tooling and infrastructure that will hopefully make the actual game development feel more like building with Legos.
The biggest development is that I re-wrote the GUI tools from tauri/typescript to egui/rust. Having a web-based UI is just too heavy and if I'm going to be using something frequently I'd want it to be quick and snappy. That in itself was challenging because the UI tooling is so much better for the web (makes sense, given that it's the dominant type of UI these days) and the Rust ecosystem in general is still in its infancy. But ultimately it was worth it.
Two completely new tools are crane and depot. On the point of wanting to avoid clunky UIs, Unity is so sluggish on Linux (not sure if the same is true for Windows) and just adds so much mental overhead to assembling scenes within it. crane is instead a very simple scene editor with modal controls (à la vi/vim; I'm a zealot) which is capable of parsing and exporting Unity scenes. So the bulk of scene editing/creation can happen in there, with some parts (MonoBehaviour scripts, etc) that might still need to be done from within Unity, but are far less common tasks.
Depot & 3d overload
depot manages all the non-character 3d assets, tracking licensing and attribution where needed (e.g. CC-BY licensed assets), and common 3d processing tasks built-in. Primarily this is generating model variations with different textures and mesh simplification (way more on this below). It'll eventually integrated with crane so crane can directly pull models from depot to add to a scene.
As I alluded to a long time ago, the 3d model aspect is probably the most difficult part of the technical aspect of the game's development (writing, character development, etc are harder—and more fun—but leaving those aside for now) and I've decided the best approach is to set aside some budget to purchase pre-existing assets and hand-create only the simplest things. The amount of time saved will be well worth the cost.
The downside of using pre-made assets, however, is a lot of variability. Meshes can vary tremendously in poly counts, textures, etc; not to mention model formats (obj, fbx, gltf/glb, etc) which is a massive challenge itself. So I basically need a processing pipeline that can take a pre-made asset in any format (or at least the major ones) and turn it into something optimized and visually consistent with everything else with as little input from me as possible. Buying pre-made assets is moot if I have to spend just as much time adjusting the model to fit my particular needs.
This led me down a dizzying rabbit hole to the world of 3d software-stuff (? not sure what to call it) and so far I've come away with the following learnings:
- There are too many 3d formats! And no reliable means of converting between all of them, or even just the major formats. There's
assimpwhich seems ill-maintained, and you can use Blender too but it's frankly heavy for the job. The best tool I've found so far istrimeshbut afaik they don't provide a command line tool or anything, so you need to have Python setup to use it. It'd be so nice to have something likeffmpegbut for 3d models. Instead I've found that specific conversion software, likeobj2gltforfbx2gltfare the most reliable. - Formats aside, there's so much variability already in meshes (welded vs unwelded, tris vs quads, messed up normals, bad UV mappings, water-tightness, etc) and thus so many opportunities for malformed meshes that will just gunk up any automated processing you have set up. Again,
trimeshhas so far seemed good at standardizing models to some degree such that they'll then work for other mesh simplification programs. - In terms of pre-made programs and libraries you often see just two extremes: extraordinarily expensive but good & robust (I'm guessing, I can't afford to try them) geared towards big money-making studios (where licenses are generally in the tens of thousands per year) and 5 to 10 year old code for a random SIGGRAPH paper that seems so promising but isn't written for general use and/or is an absolute nightmare to compile. There are things in between but they're relatively rare. In contrast, for data science, for example, there is a massive wealth of libraries/software of different scales out there to use.
- Related to the previous point, I've found that where native Rust libraries exist for some of these they're often unexpectedly lopsided. For example, the
tobjcrate can load.objs but it can't save them. I don't know enough about the format to say more—I'm sure there's a good reason, but it just seems strange to me. - The ecosystem is still very Windows-focused, given that Windows is still the dominant computer gaming platform.
wineis a godsend here, at least for running.exes; so far I haven't had much luck compiling Windows C++ programs (usingmsbuildormsvcorclangor whatever).
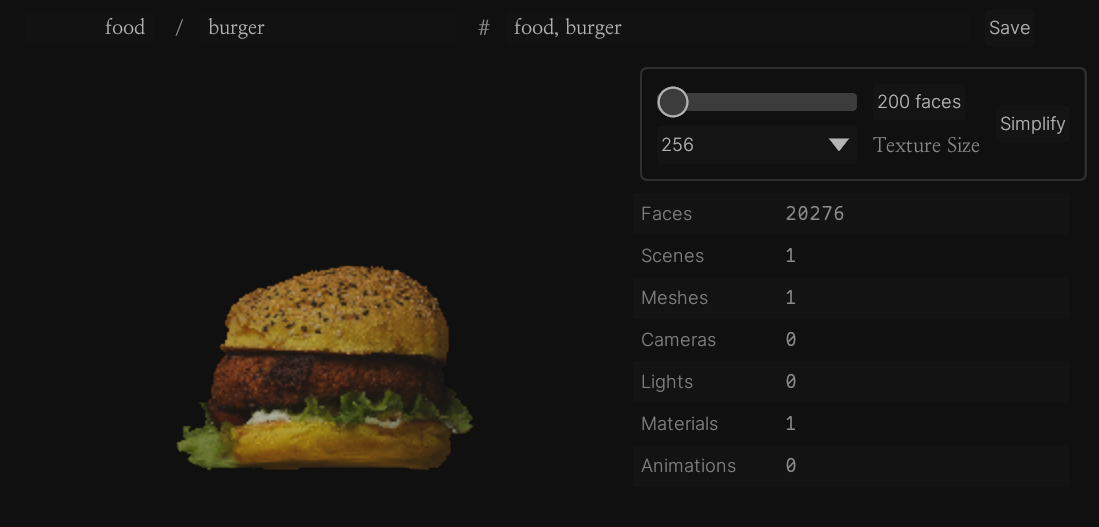
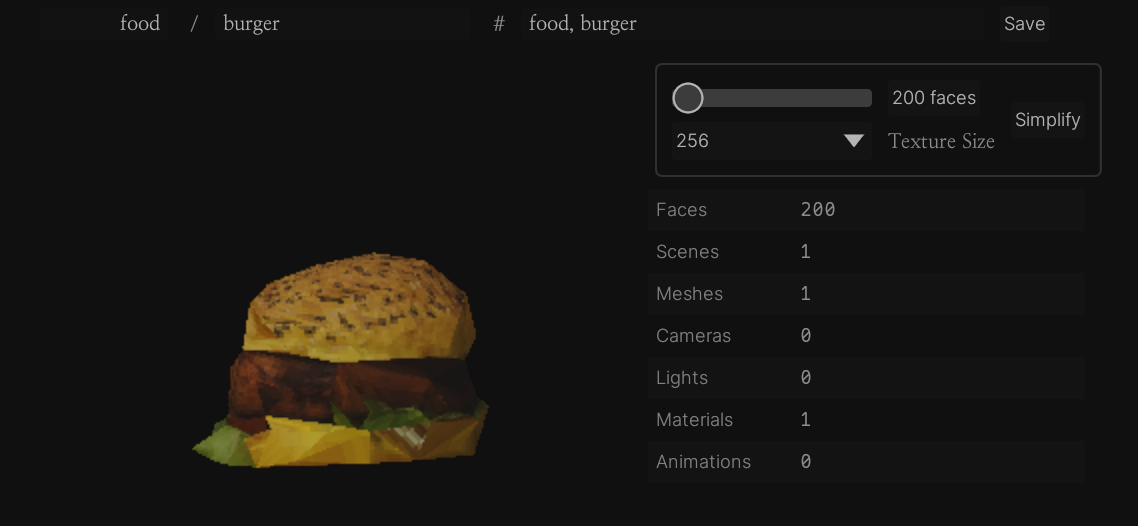
Reducing polys
There are a lot of different 3d model processing tasks but fortunately for me the only one I'm concerned with is "mesh simplification", i.e. taking a high-poly mesh and turning it into a low-poly one that still resembles the original. Generally you will bake some of the high-poly mesh information as textures (e.g. a normal map) to help with this.
There's a related task of creating an "imposter", where we don't modify the mesh itself but instead create a simple proxy polygon, e.g. an octahedron. You then pre-render the object from various perspectives and those are then projected onto the polygon. The idea here is if an object is going to be static and only viewed from a few angles, we don't need to store/render all the geometry as we're essentially looking at a 2d image.
Imposters are probably the right approach for set-piece elements and the like. They're much easier for me conceptually and because of their simplicity are probably going to be more robust than actually mutating the mesh. There are plenty of Unity plugins that do this; I haven't tried them yet but they're all well-reviewed.
But for anything more complicated, especially things that need to move, imposters won't be enough. We have to actually change the geometry to reduce the poly count.
I've seen a few different terms for this task; I'm not sure what the distinctions are between them, if any: simplification, decimation, remeshing, retopology, reduction, LOD generation, and probably more.
Reflecting the pattern above there are a few "pro" options for this task, namely Simplygon which seems to be the industry standard (and if I'm not mistaken is integrated into the Unreal Engine's mesh simplification routines), evident in its whopping cost of $35,000 per year. They do have a free version but it's very limited. There's also InstaLOD which has a more generous free option, but I haven't been able to download it and I think it excludes the use of the C++ API. When there are free offerings direct API access is usually behind the paywall or, when no free offering exist, behind the more expensive plan. For example, with Pixyz it's $1,350/year for just the plugin and $2,450/year for the Python API.
So I haven't tried any of these products but I wonder how much better they are in terms of raw mesh simplification quality. They probably have a bunch of other features important to giant studios like build system integration or support for more esoteric workflows/needs; that might be more of the selling point than how good their mesh simplification algorithm is. I wonder this partly because after looking through the free or cheaper options they mostly use the same approach: some variation of quadric mesh simplification. As I understand this basically involves computing a metric (the quadric metric) for each edge in the mesh, essentially ranking the edges according to how important they are (I'm waving my hands) and then deleting the least important edge. Rinse and repeat until you hit your target face/vert count.
Some examples that I believe use this approach are SeamAwareDecimater, FastQuadricMeshSimplification, MeshLab, and nanomesh. On my test meshes they all give similar results. It feels like this approach is better suited to preserving as much detail as possible when simplifying, which is great for going from high-poly to mid-poly, but when you start to get to the very low poly ranges (<3000 faces or so) the mesh just starts to degrade, have holes, etc.
This thread discusses some of the other shortcomings of this approach and is where I first encountered the term "silhouette protection". With the quadric approach you eventually get to the point where you start deleting faces and start to lose essential elements of the model, until it becomes unrecognizable. Silhouette protection is meant to stop the algorithm before it reaches this point; or it may be a totally different simplification approach, I don't really know.
One approach I thought might work is to take the convex hull of the mesh and iteratively introduce edges, essentially "shrink-wrapping" the hull around the original mesh until you reach the desired number of faces or some minimum error between the mesh and the hull. The downside with this approach is that I don't think it'd work for meshes that have holes because a convex hull doesn't take those into account (afaik).
I wasn't able to find much pre-existing software that does this, just Semi Convex Hull for Mesh Simplification and Boundingmesh which uses "bounding convex decomposition". I wasn't able to get them to finish simplifying my heavier test meshes, they ran without terminating. However, I believe this approach is kind of how collider meshes are computed, but looking at this collider mesh generation plugin it seems like the approach (for at least the VHACD algorithm; see also and here) is to break down the mesh into different parts and then build the collider mesh from the convex hulls of those individual parts, thus the mesh is no longer contiguous. But this does get around the hole problem I mentioned, and I guess you could re-join the individual parts though.
So unfortunately no clear resolution yet, but there's still more to try.